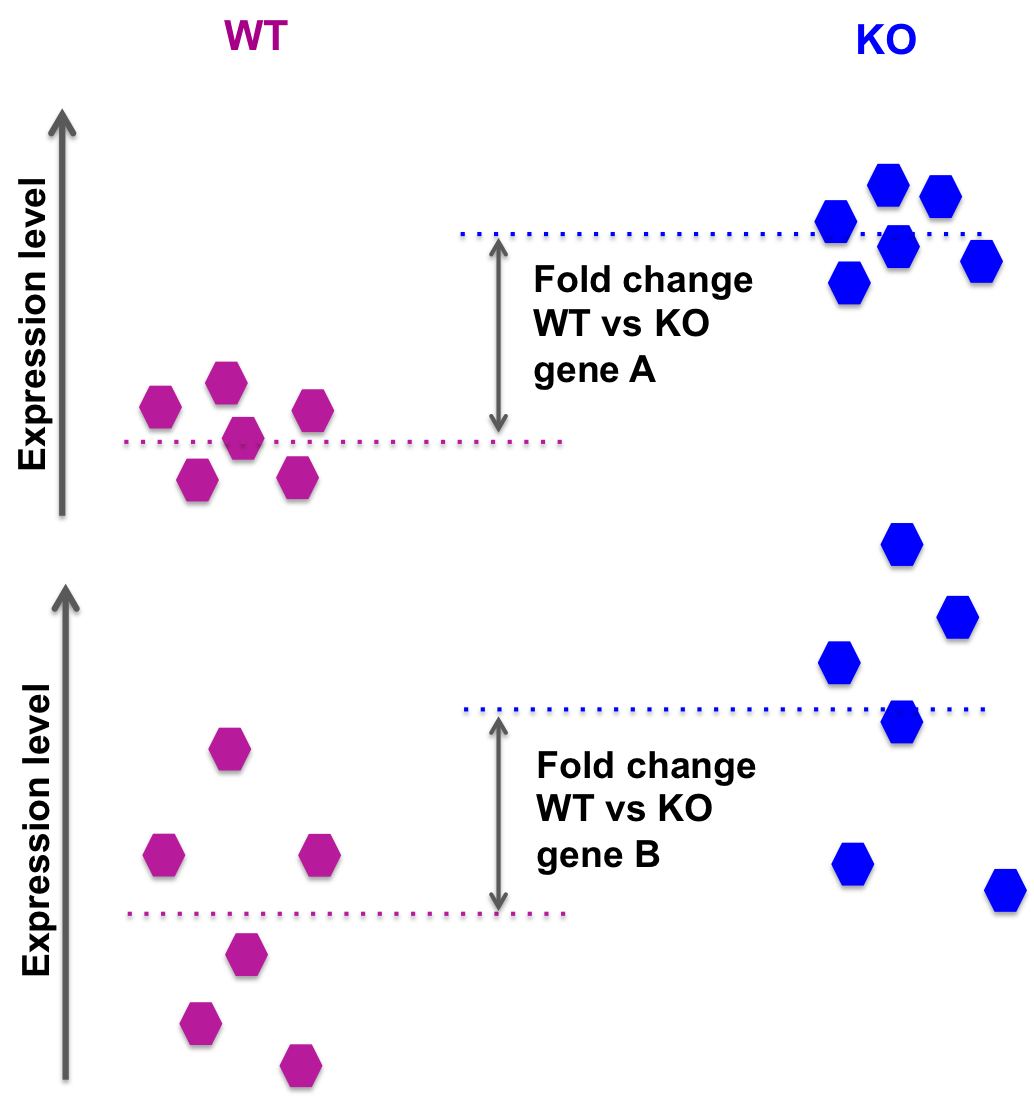
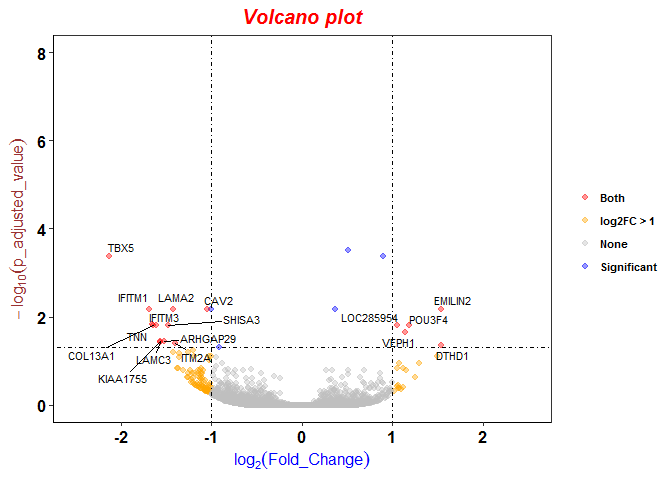
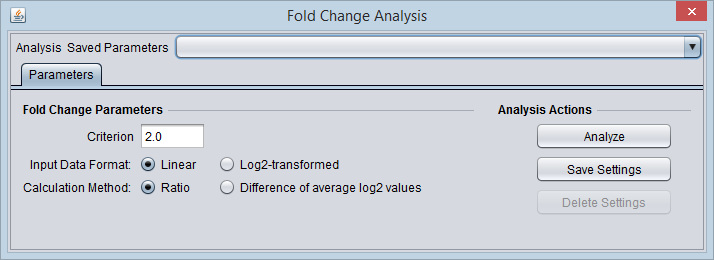
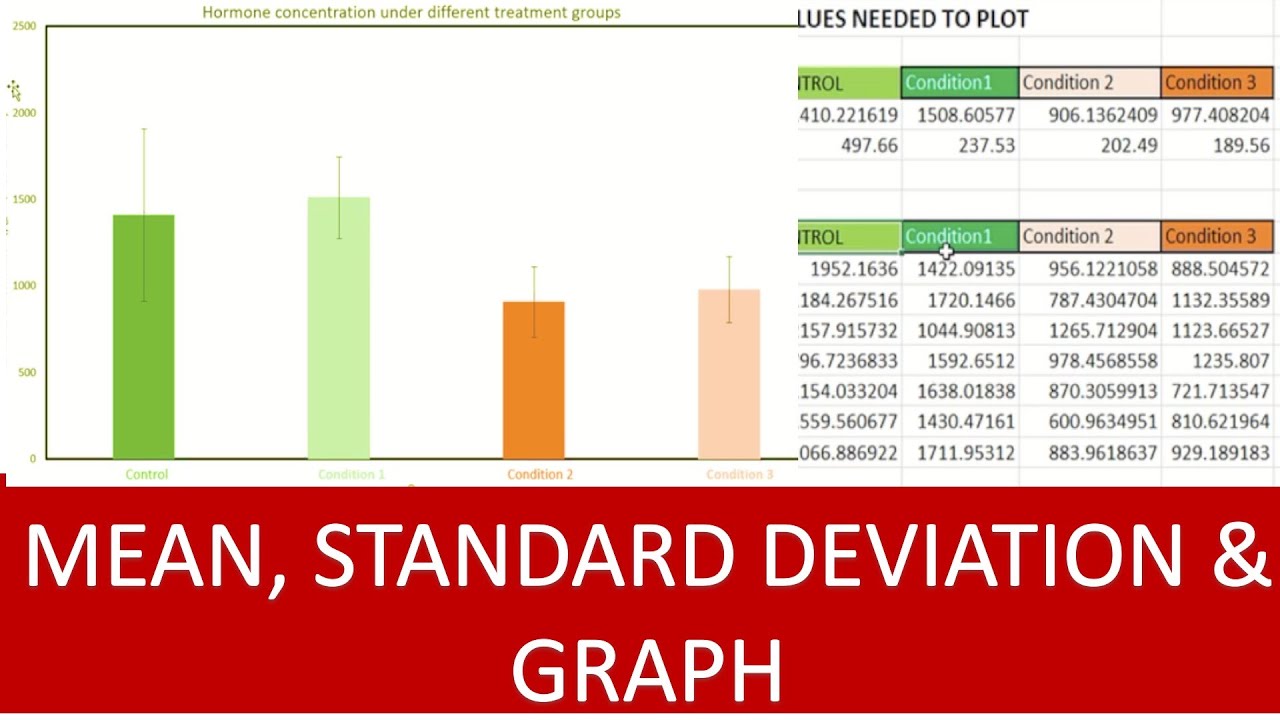
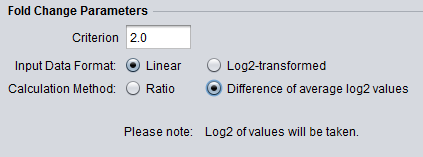
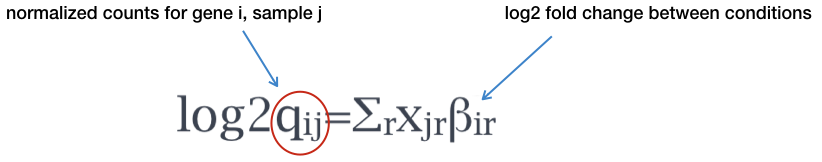Different logFC (log2foldchange) values for genes from limma-voom and other tools (edgeR and DESeq2)

PCRdDCq.png. Relative (log2) fold change between case and control for a... | Download Scientific Diagram
Log2 fold changes in rank_genes_groups are calculated from log-transformed data · Issue #517 · scverse/scanpy · GitHub
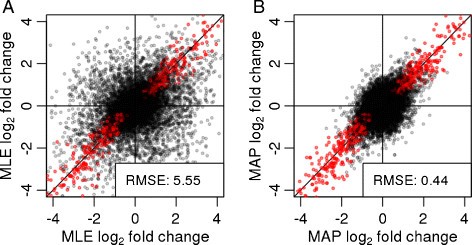
Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2 | Genome Biology | Full Text

The distribution of log2 fold changes when the expression levels from... | Download Scientific Diagram

Linear regression analysis of fold changes calculated from qPCR and... | Download Scientific Diagram